
Atomistic Simulations and In Silico Mutational Profiling of Protein Stability and Binding in the SARS-CoV-2 Spike Protein Complexes with Nanobodies: Molecular Determinants of Mutational Escape Mechanisms
4.7
(123) ·
$ 4.99 ·
In stock
Description

Impact of new variants on SARS-CoV-2 infectivity and neutralization: A molecular assessment of the alterations in the spike-host protein interactions - ScienceDirect

Versatile and multivalent nanobodies efficiently neutralize SARS-CoV-2

Mutational landscape and in silico structure models of SARS-CoV-2 spike receptor binding domain reveal key molecular determinants for virus-host interaction, BMC Molecular and Cell Biology
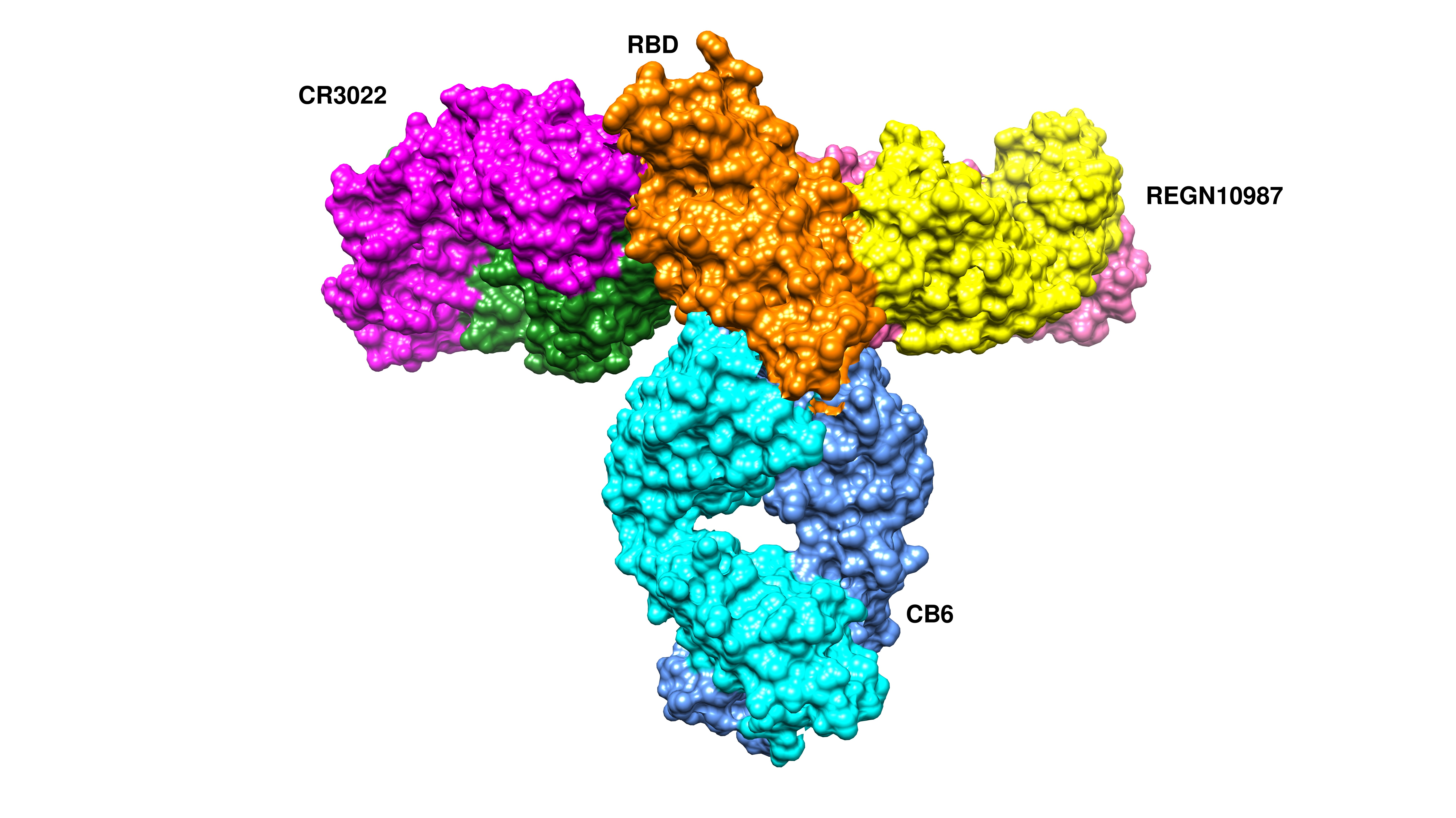
Cells, Free Full-Text

Machine learning and protein allostery: Trends in Biochemical Sciences

A tethered ligand assay to probe SARS-CoV-2:ACE2 interactions
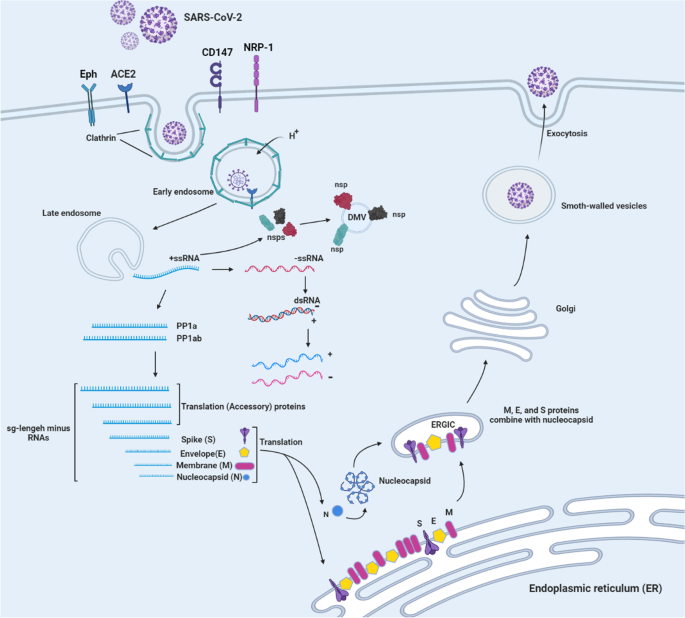
Structural and non-structural proteins in SARS-CoV-2: potential aspects to COVID-19 treatment or prevention of progression of related diseases, Cell Communication and Signaling
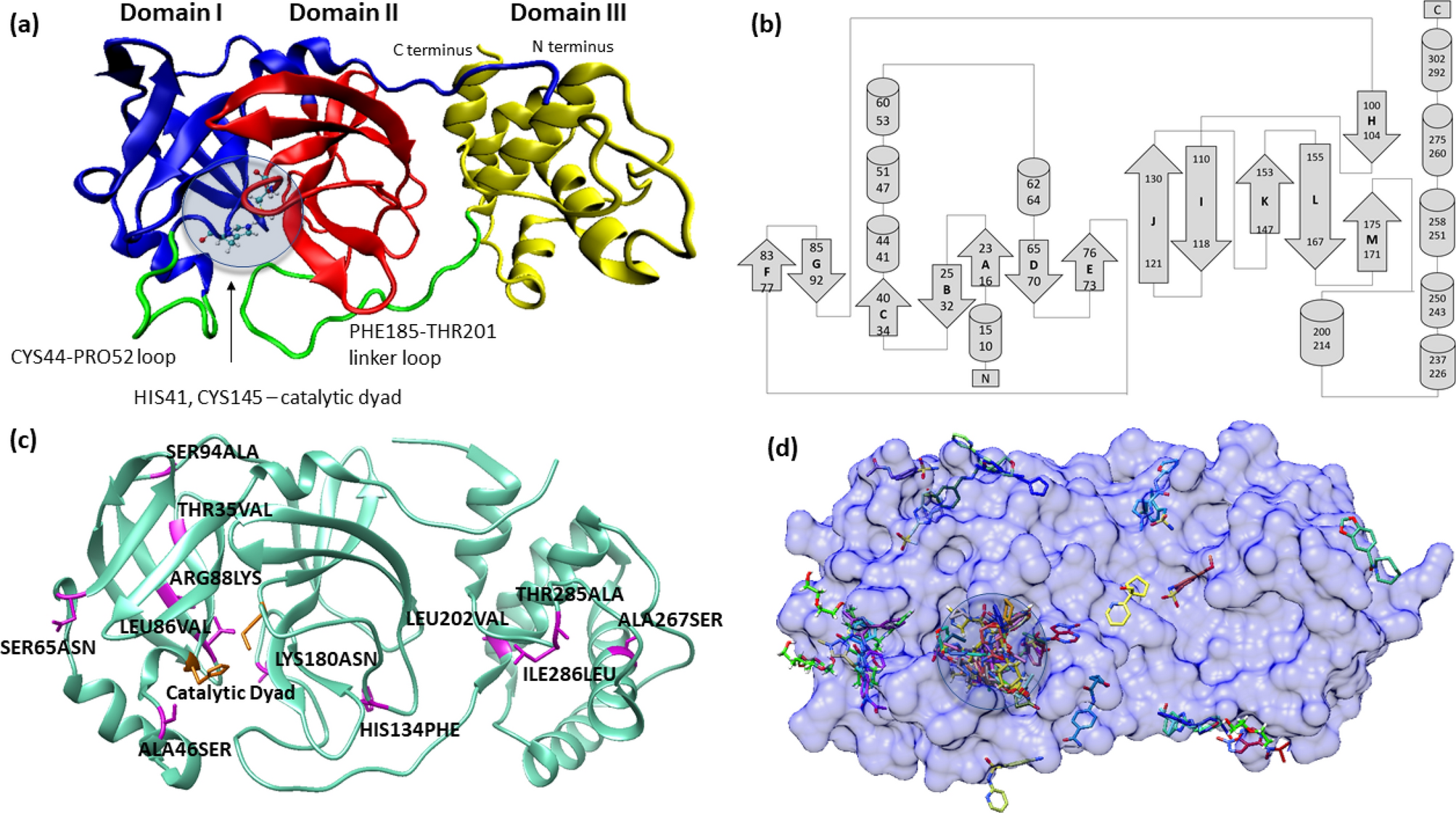
Molecular dynamics and in silico mutagenesis on the reversible inhibitor-bound SARS-CoV-2 main protease complexes reveal the role of lateral pocket in enhancing the ligand affinity

Mutational landscape and in silico structure models of SARS-CoV-2 spike receptor binding domain reveal key molecular determinants for virus-host interaction, BMC Molecular and Cell Biology
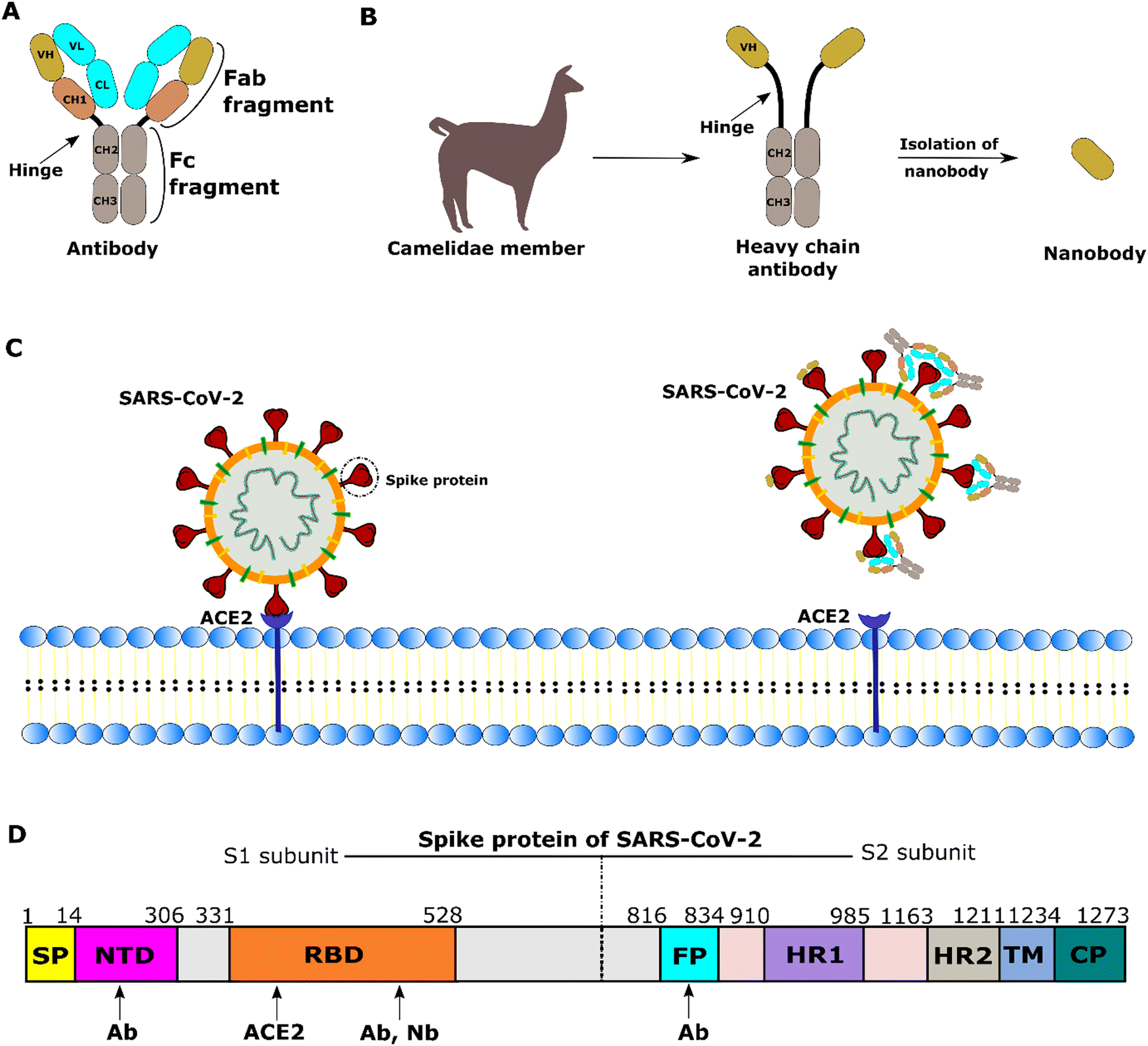
Interaction of SARS-CoV-2 with host cells and antibodies: experiment and simulation - Chemical Society Reviews (RSC Publishing) DOI:10.1039/D1CS01170G
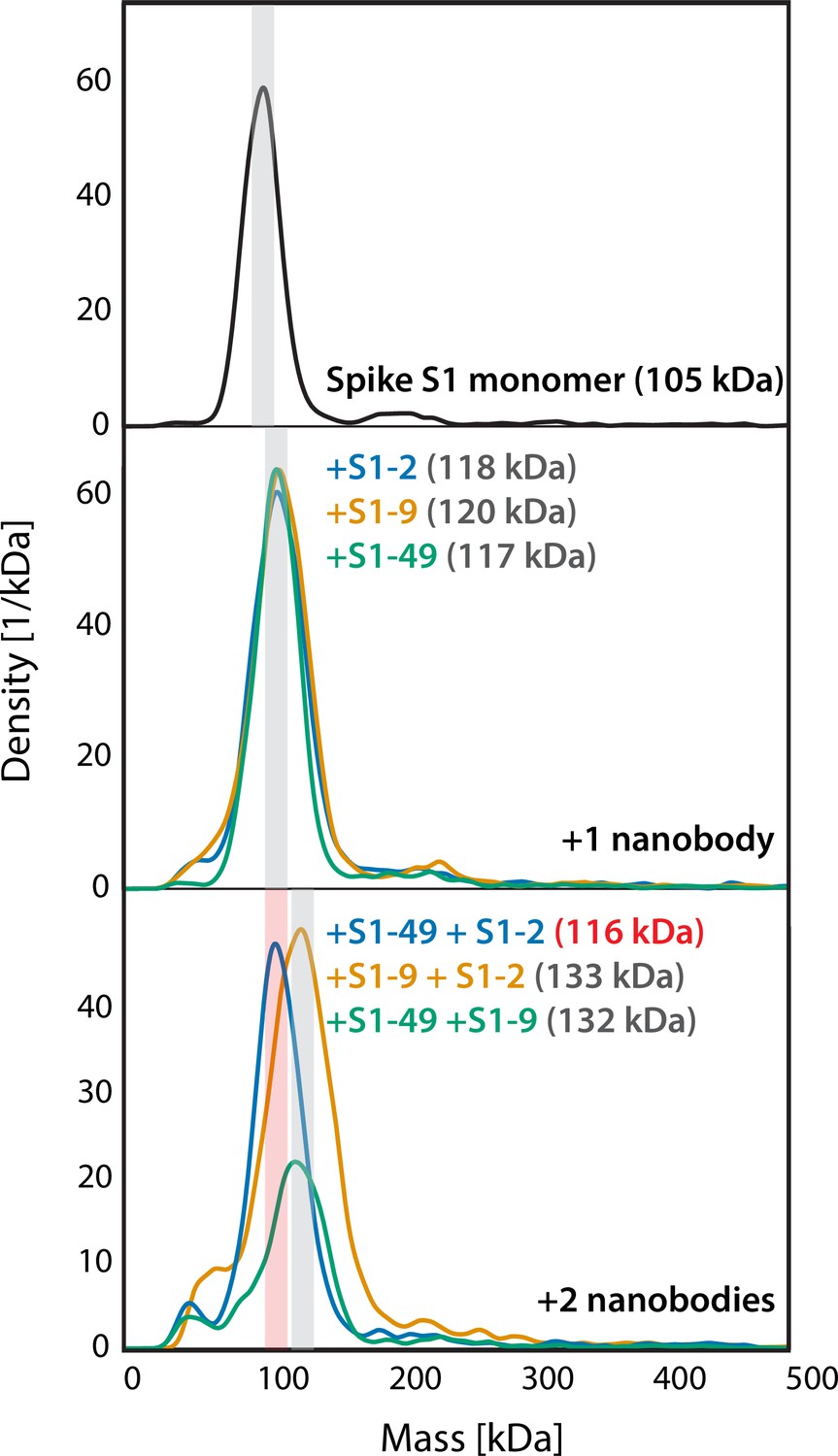
Highly synergistic combinations of nanobodies that target SARS-CoV-2 and are resistant to escape

Highly synergistic combinations of nanobodies that target SARS-CoV-2 and are resistant to escape
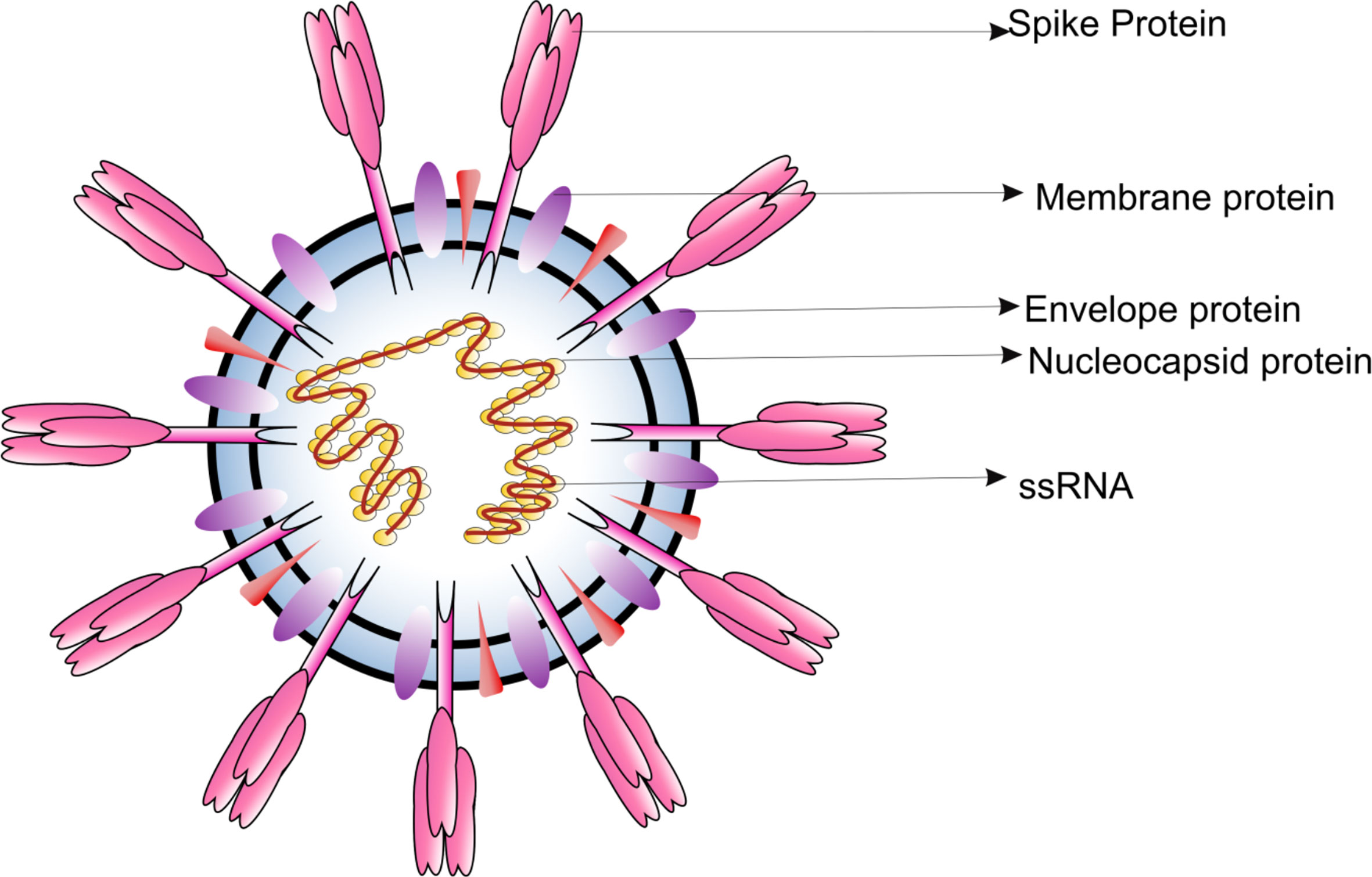
Frontiers Potential Therapeutic Targets and Vaccine Development for SARS- CoV-2/COVID-19 Pandemic Management: A Review on the Recent Update
Related products
You may also like
copyright © 2019-2024 tlpa.aero all rights reserved.








